Plotting Bayesian Analysis#
A number of functions exist for plotting the results of Bayesian analysis. The problem and result used in this section were made using the
DSPC Standard Layers example and a control object with the procedure set to DREAM.
% Run the DSPC standard layers example
controls = controlsClass();
controls.procedure = 'dream';
[problem, results] = RAT(problem, controls);
# Run the DSPC standard layer example
controls = RAT.Controls()
controls.procedure = 'dream'
problem, results = RAT.run(problem, controls)
Reflectivity and SLD#
A simple reflectivity shaded plot can be displayed as follows:
bayesShadedPlot(problem, results)
RAT.plotting.plot_ref_sld(problem, results, bayes=65)
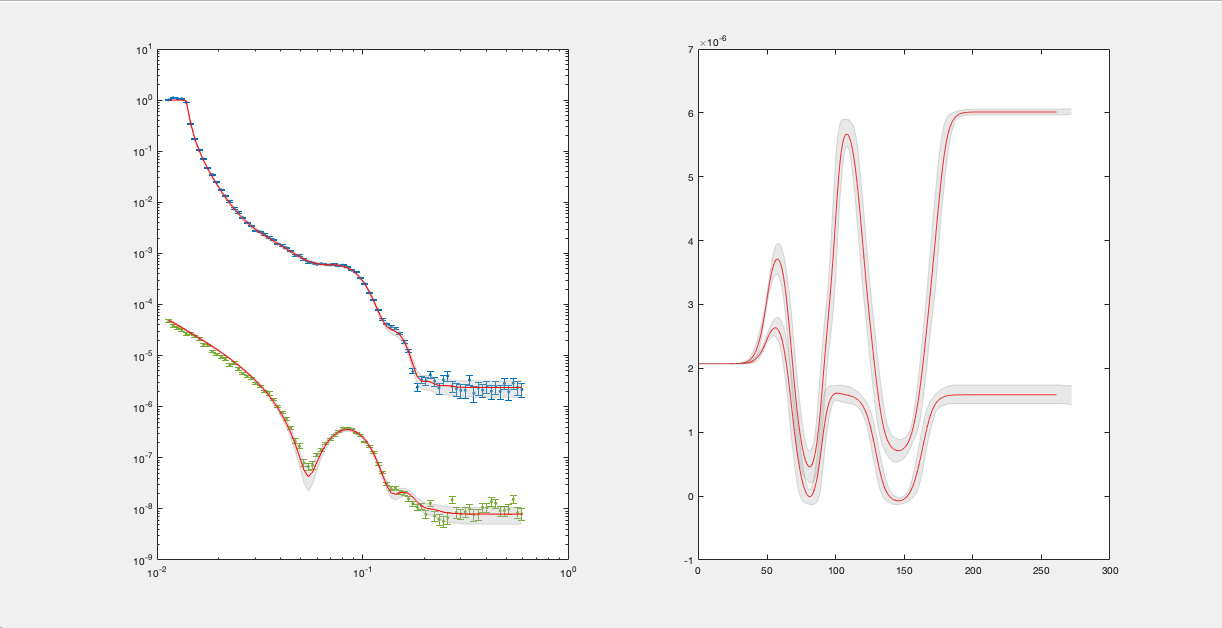
By default, this shows a standard reflectivity plot with a 65% shaded confidence interval.
There are a number of options to customise the plot:
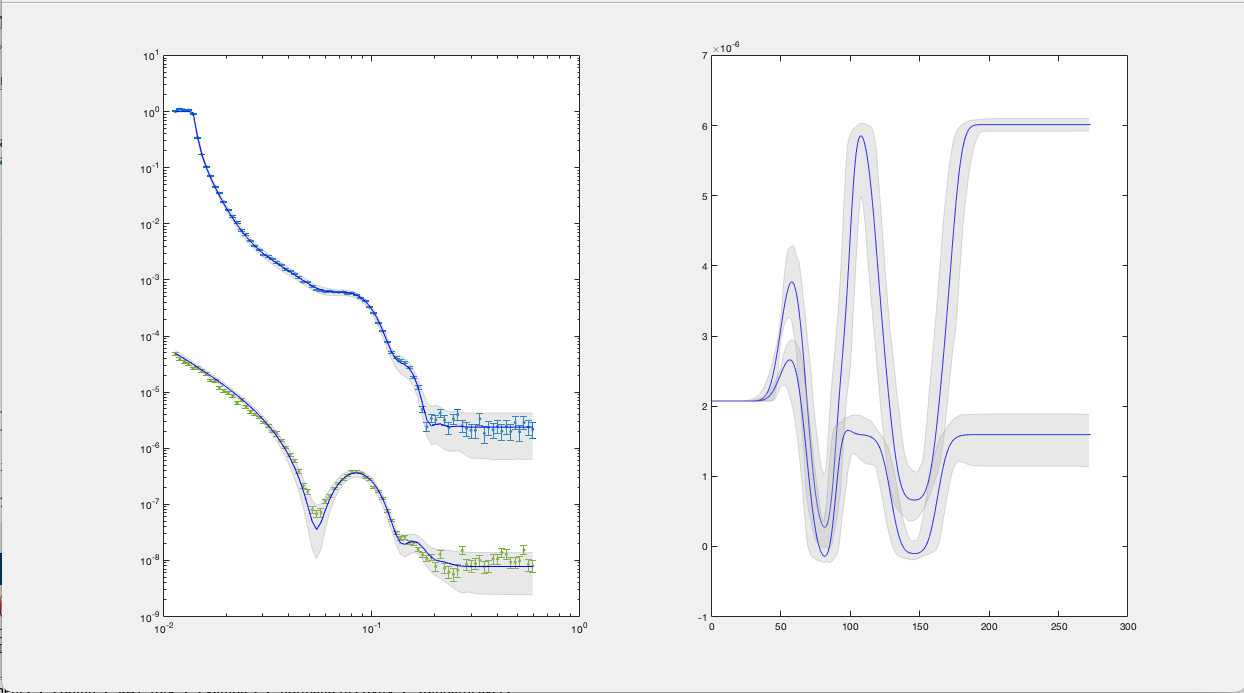
Interval - You can sepcify either the 65% or 95% confidence interval to display:
bayesShadedPlot(problem, results, 'interval', 95)
RAT.plotting.plot_ref_sld(problem, results, bayes=95)
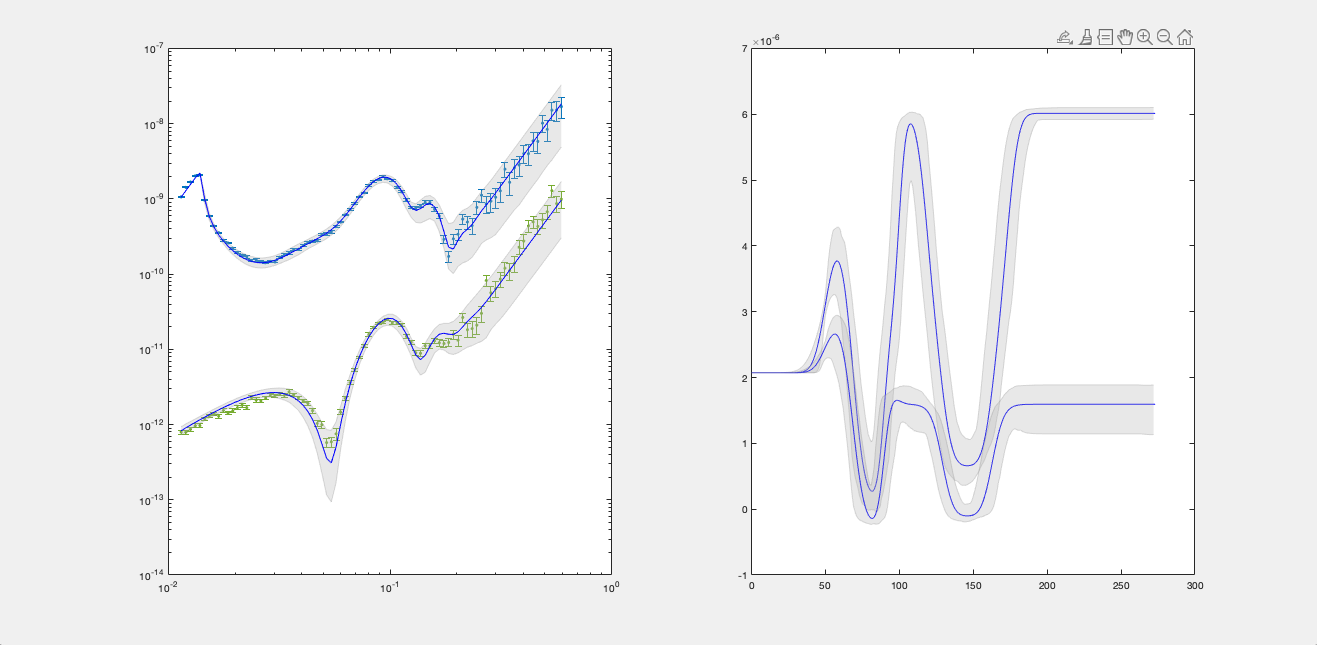
Type - You can also specify a q4 plot for the reflectivity:
bayesShadedPlot(problem, results, 'q4', true)
RAT.plotting.plot_ref_sld(problem, results, bayes=65, q4=True)
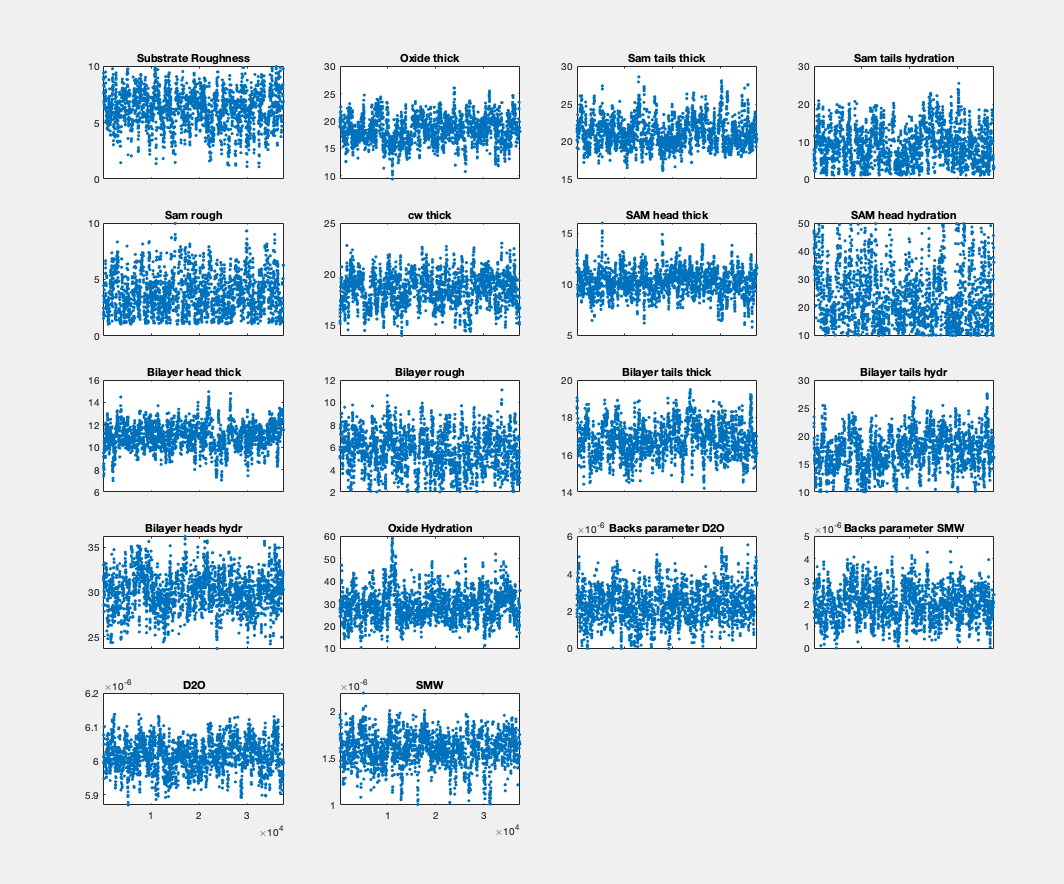
Posterior Histograms#
You can easily view the marginalised Bayesian posteriors from your analysis:
plotHists(results)
RAT.plotting.plot_hists(results)
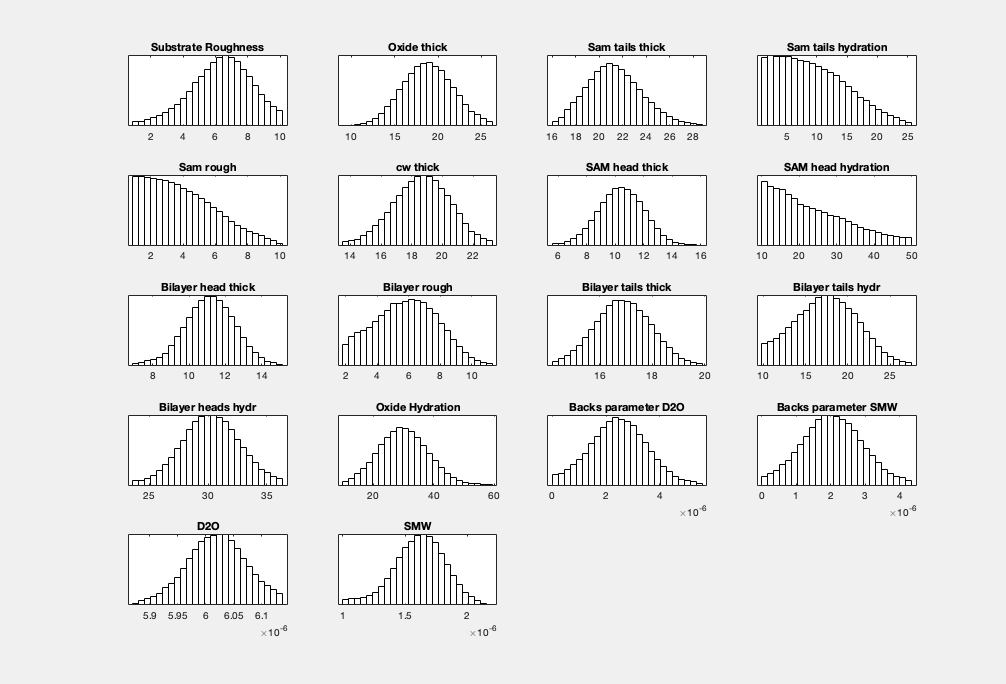
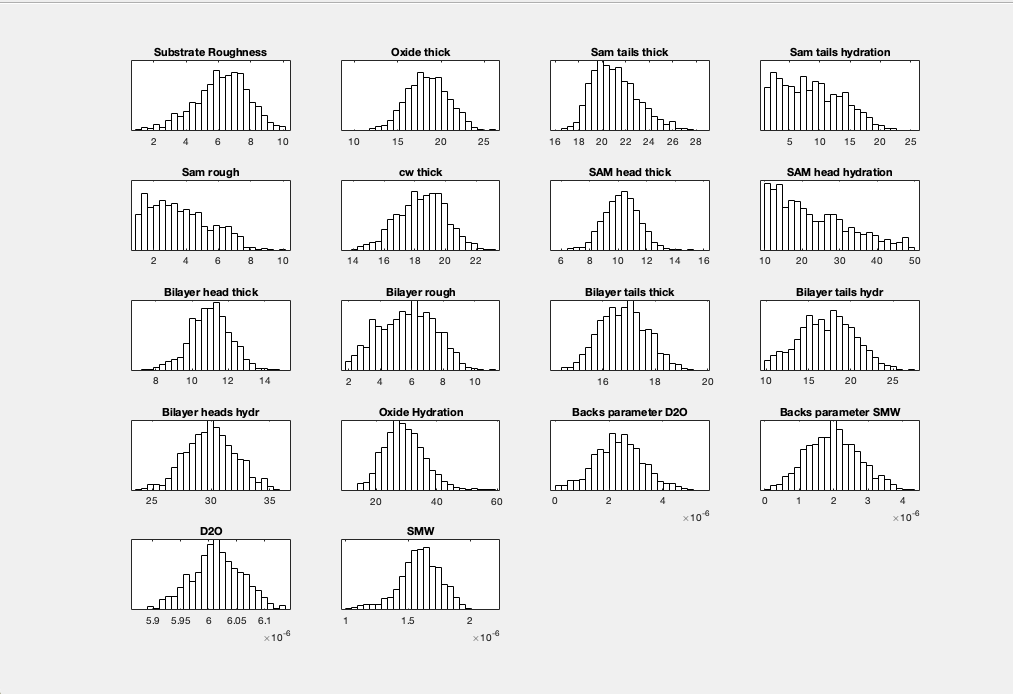
By default, plotHists carries out a KDE smooth of the histograms. You can optionally choose no smoothing:
plotHists(results,'smooth',false)
RAT.plotting.plot_hists(results, smooth=False)
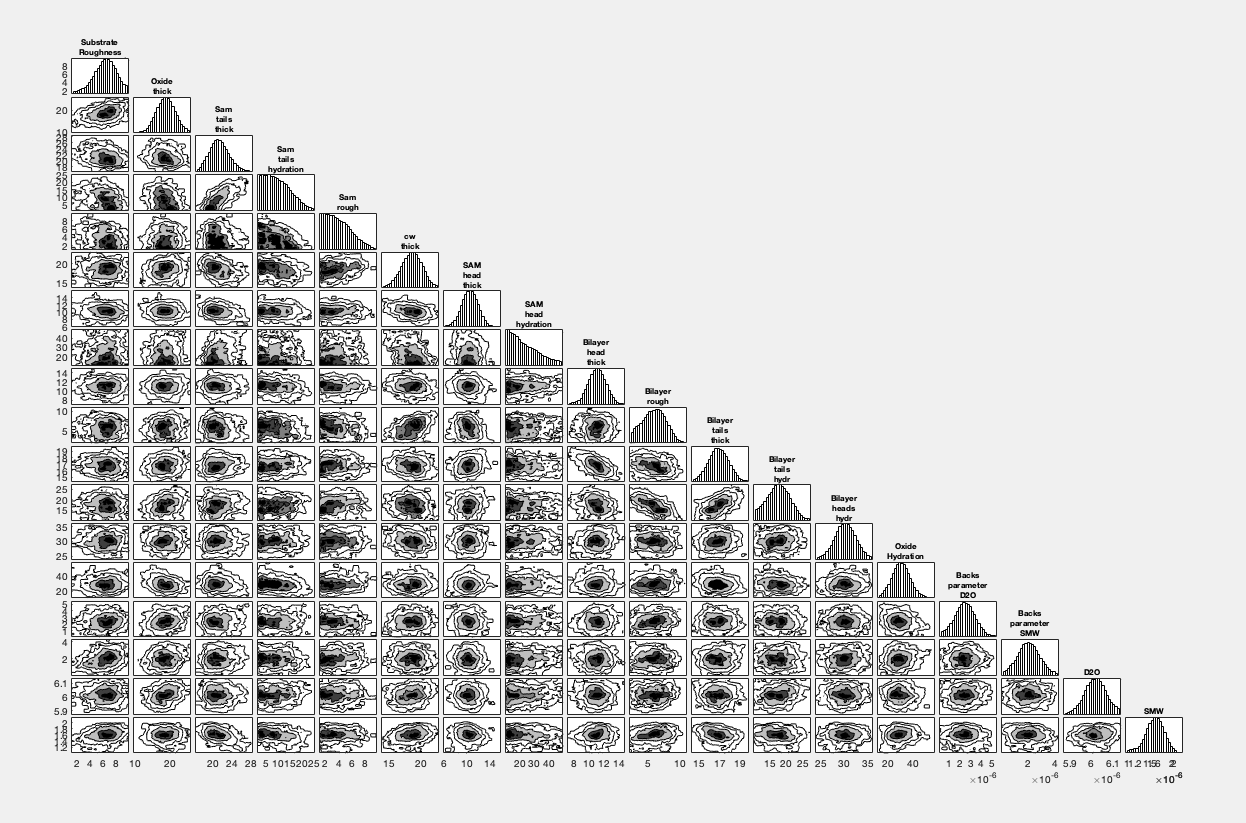
Corner Plots#
To produce a cornerplot, simply use the cornerPlot function:
cornerPlot(results)
RAT.plotting.plot_corner(results)
Chain View#
Finally, you can check the integrity of your markov chain as follows:
plotChain(results);
RAT.plotting.plot_chain(results)