Note
To get the output project and results from this example in your Python session, run:
from ratapi.examples import absorption
project, results = absorption()
[1]:
import pathlib
import numpy as np
from IPython.display import Code
import ratapi as RAT
from ratapi.models import Parameter
Absorption (imaginary SLD) - effect below the critical edge#
RAT allows the use of an imaginary, as well as real part of the SLD. The effect of this is usually seen below the critical edge, and must sometimes be accounted for.
The example used here is Custom Layers. It analyses a bilayer sample on a permalloy / gold substrate, measured using polarised neutrons, against D2O and H2O, leading to 4 contrasts in total. Absorption (i.e. imaginary SLD) is defined for Gold and the Permalloy, to account for non-flat data below the critical edge.
For absorption with standard layers, an additional column appears in the layers block to accommodate the imagainary component of the SLD. For custom functions, we add an extra column to the output.
For all calculation types, to activate this functionality it is necessary to set the ‘absorption’ flag when creating the project.
[2]:
problem = RAT.Project(name="Absorption example", calculation="normal", model="custom layers", geometry="substrate/liquid", absorption=True)
We now define our parameters, noting that each SLD parameter has both a real and imaginary component:
[3]:
parameter_list = [
Parameter(name="Alloy Thickness", min=100.0, value=135.6, max=200.0, fit=True),
Parameter(name="Alloy SLD up", min=6.0e-6, value=9.87e-6, max=1.2e-5, fit=True),
Parameter(name="Alloy SLD imaginary up", min=1.0e-9, value=4.87e-8, max=1.0e-7, fit=True),
Parameter(name="Alloy SLD down", min=6.0e-6, value=7.05e-6, max=1.3e-5, fit=True),
Parameter(name="Alloy SLD imaginary down", min=1.0e-9, value=4.87e-8, max=1.0e-7, fit=True),
Parameter(name="Alloy Roughness", min=2.0, value=5.71, max=10.0, fit=True),
#
Parameter(name="Gold Thickness", min=100.0, value=154.7, max=200.0, fit=True),
Parameter(name="Gold Roughness", min=0.1, value=5.42, max=10.0, fit=True),
Parameter(name="Gold SLD", min=4.0e-6, value=4.49e-6, max=5.0e-6, fit=True),
Parameter(name="Gold SLD imaginary", min=1.0e-9, value=4.20e-8, max=1.0e-7, fit=True),
#
Parameter(name="Thiol APM", min=40.0, value=56.27, max=100.0, fit=True),
Parameter(name="Thiol Head Hydration", min=20.0, value=30.0, max=50.0, fit=True),
Parameter(name="Thiol Coverage", min=0.5, value=0.9, max=1.0, fit=True),
#
Parameter(name="CW Thickness", min=1.0, value=12.87, max=25.0, fit=True),
#
Parameter(name="Bilayer APM", min=48.0, value=65.86, max=90.0, fit=True),
Parameter(name="Bilayer Head Hydration", min=20.0, value=30.0, max=50.0, fit=True),
Parameter(name="Bilayer Roughness", min=1.0, value=3.87, max=10.0, fit=True),
Parameter(name="Bilayer Coverage", min=0.5, value=0.94, max=1.0, fit=True)
]
problem.parameters.extend(parameter_list)
Set the bulk in and bulk out parameters:
[4]:
problem.bulk_in.set_fields(0, name="Silicon", min=2.0e-6, value=2.073e-6, max=2.1e-6)
problem.bulk_out.set_fields(0, name="D2O", min=5.8e-06, value=6.21e-06, max=6.35e-06, fit=True)
problem.bulk_out.append(name="H2O", min=-5.6e-07, value=-3.15e-07, max=0.0, fit=True)
Use a different scalefactor for each dataset:
[5]:
del problem.scalefactors[0]
problem.scalefactors.append(name="Scalefactor 1", min=0.5, value=1, max=1.5, fit=True)
problem.scalefactors.append(name="Scalefactor 2", min=0.5, value=1, max=1.5, fit=True)
problem.scalefactors.append(name="Scalefactor 3", min=0.5, value=1, max=1.5, fit=True)
problem.scalefactors.append(name="Scalefactor 4", min=0.5, value=1, max=1.5, fit=True)
Set the backgrounds and resolutions:
[6]:
del problem.backgrounds[0]
del problem.background_parameters[0]
problem.background_parameters.append(name="Background parameter 1", min=5.0e-08, value=7.88e-06, max=9.0e-05, fit=True)
problem.background_parameters.append(name="Background parameter 2", min=1.0e-08, value=5.46e-06, max=9.0e-05, fit=True)
problem.background_parameters.append(name="Background parameter 3", min=1.0e-06, value=9.01e-06, max=9.0e-05, fit=True)
problem.background_parameters.append(name="Background parameter 4", min=1.0e-06, value=5.61e-06, max=9.0e-05, fit=True)
problem.backgrounds.append(name="Background 1", type="constant", source="Background parameter 1")
problem.backgrounds.append(name="Background 2", type="constant", source="Background parameter 2")
problem.backgrounds.append(name="Background 3", type="constant", source="Background parameter 3")
problem.backgrounds.append(name="Background 4", type="constant", source="Background parameter 4")
# Make the resolution fittable
problem.resolution_parameters.set_fields(0, fit=True)
Add the datasets:
[7]:
data_path = pathlib.Path("../data")
data_1 = np.loadtxt(data_path / "D2O_spin_down.dat")
problem.data.append(name="D2O_dn", data=data_1)
data_2 = np.loadtxt(data_path / "D2O_spin_up.dat")
problem.data.append(name="D2O_up", data=data_2)
data_3 = np.loadtxt(data_path / "H2O_spin_down.dat")
problem.data.append(name="H2O_dn", data=data_3)
data_4 = np.loadtxt(data_path / "H2O_spin_up.dat")
problem.data.append(name="H2O_up", data=data_4)
Add the custom file. We can see that we add an extra column for the output in our custom function.
[8]:
problem.custom_files.append(
name="DPPC absorption",
filename="volume_thiol_bilayer.py",
language="python",
path=pathlib.Path.cwd().resolve(),
)
Code(filename='volume_thiol_bilayer.py', language='python')
[8]:
"""Custom layer model file for the absorption example."""
def volume_thiol_bilayer(params, bulk_in, bulk_out, contrast):
"""VolumeThiolBilayer RAT Custom Layer Model File.
This file accepts 3 vectors containing the values for params, bulk in and bulk out.
The final parameter is an index of the contrast being calculated
The function should output a matrix of layer values, in the form...
Output = [thick 1, SLD 1, Rough 1, Percent Hydration 1, Hydrate how 1
....
thick n, SLD n, Rough n, Percent Hydration n, Hydration how n]
The "hydrate how" parameter decides if the layer is hydrated with Bulk out or Bulk in phases.
Set to 1 for Bulk out, zero for Bulk in.
Alternatively, leave out hydration and just return...
Output = [thick 1, SLD 1, Rough 1,
....
thick n, SLD n, Rough n]
The second output parameter should be the substrate roughness.
"""
# Note - The first contrast number is 1 (not 0) so be careful if you use
# this variable for array indexing.
subRough = params[0]
alloyThick = params[1]
alloySLDUp = params[2]
alloyISLDUp = params[3]
alloySLDDown = params[4]
alloyISLDDown = params[5]
alloyRough = params[6]
goldThick = params[7]
goldRough = params[8]
goldSLD = params[9]
goldISLD = params[10]
thiolAPM = params[11]
thiolHeadHydr = params[12]
thiolCoverage = params[13]
cwThick = params[14]
bilayerAPM = params[15]
bilHeadHydr = params[16]
bilayerRough = params[17]
bilayerCoverage = params[18]
# Make the metal layers
gold = [goldThick, goldSLD, goldISLD, goldRough]
alloyUp = [alloyThick, alloySLDUp, alloyISLDUp, alloyRough]
alloyDown = [alloyThick, alloySLDDown, alloyISLDDown, alloyRough]
# Neutron b's..
# define all the neutron b's.
bc = 0.6646e-4 # Carbon
bo = 0.5843e-4 # Oxygen
bh = -0.3739e-4 # Hydrogen
bp = 0.513e-4 # Phosphorus
bn = 0.936e-4 # Nitrogen
# Work out the total scattering length in each fragment
# Define scattering lengths
# Hydrogenated version
COO = (2 * bo) + (bc)
GLYC = (3 * bc) + (5 * bh)
CH3 = (1 * bc) + (3 * bh)
PO4 = (1 * bp) + (4 * bo)
CH2 = (1 * bc) + (2 * bh)
CH = (1 * bc) + (1 * bh)
CHOL = (5 * bc) + (12 * bh) + (1 * bn)
# And also volumes
vCH3 = 52.7 # CH3 volume in the paper appears to be for 2 * CH3's
vCH2 = 28.1
vCOO = 39.0
vGLYC = 68.8
vPO4 = 53.7
vCHOL = 120.4
vCHCH = 42.14
vHead = vCHOL + vPO4 + vGLYC + 2 * vCOO
vTail = (28 * vCH2) + (1 * vCHCH) + (2 * vCH3) # Tail volume
# Calculate sum_b's for other fragments
sumbHead = CHOL + PO4 + GLYC + 2 * COO
sumbTail = (28 * CH2) + (2 * CH) + 2 * CH3
# Calculate SLDs and Thickness
sldHead = sumbHead / vHead
thickHead = vHead / thiolAPM
sldTail = sumbTail / vTail
thickTail = vTail / thiolAPM
# Correct head SLD based on hydration
thiolHeadHydr = thiolHeadHydr / 100
sldHead = sldHead * (1 - thiolHeadHydr) + (thiolHeadHydr * bulk_out[contrast - 1])
# Now correct both the SLDs for the coverage parameter
sldTail = (thiolCoverage * sldTail) + ((1 - thiolCoverage) * bulk_out[contrast - 1])
sldHead = (thiolCoverage * sldHead) + ((1 - thiolCoverage) * bulk_out[contrast - 1])
SAMTAILS = [thickTail, sldTail, 0, goldRough]
SAMHEAD = [thickHead, sldHead, 0, goldRough]
# Now do the same for the bilayer
vHead = vCHOL + vPO4 + vGLYC + 2 * vCOO
vTail = 28 * vCH2 # Tail volume
vMe = 2 * vCH3
sumbHead = CHOL + PO4 + GLYC + 2 * COO
sumbTail = 28 * CH2
sumbMe = 2 * CH3
sldHead = sumbHead / vHead
thickHead = vHead / bilayerAPM
bilHeadHydr = bilHeadHydr / 100
sldHead = sldHead * (1 - bilHeadHydr) + (bilHeadHydr * bulk_out[contrast - 1])
sldTail = sumbTail / vTail
thickTail = vTail / bilayerAPM
sldMe = sumbMe / vMe
thickMe = vMe / bilayerAPM
sldTail = (bilayerCoverage * sldTail) + ((1 - bilayerCoverage) * bulk_out[contrast - 1])
sldHead = (bilayerCoverage * sldHead) + ((1 - bilayerCoverage) * bulk_out[contrast - 1])
sldMe = (bilayerCoverage * sldMe) + ((1 - bilayerCoverage) * bulk_out[contrast - 1])
BILTAILS = [thickTail, sldTail, 0, bilayerRough]
BILHEAD = [thickHead, sldHead, 0, bilayerRough]
BILME = [thickMe, sldMe, 0, bilayerRough]
BILAYER = [BILHEAD, BILTAILS, BILME, BILME, BILTAILS, BILHEAD]
CW = [cwThick, bulk_out[contrast - 1], 0, bilayerRough]
if contrast == 2 or contrast == 4:
output = [alloyUp, gold, SAMTAILS, SAMHEAD, CW, *BILAYER]
elif contrast == 1 or contrast == 3:
output = [alloyDown, gold, SAMTAILS, SAMHEAD, CW, *BILAYER]
return output, subRough
Finally, add the contrasts:
[9]:
problem.contrasts.append(
name="D2O Down",
data="D2O_dn",
background="Background 1",
bulk_in="Silicon",
bulk_out="D2O",
scalefactor="Scalefactor 1",
resolution="Resolution 1",
resample=True,
model=["DPPC absorption"],
)
problem.contrasts.append(
name="D2O Up",
data="D2O_up",
background="Background 2",
bulk_in="Silicon",
bulk_out="D2O",
scalefactor="Scalefactor 2",
resolution="Resolution 1",
resample=True,
model=["DPPC absorption"],
)
problem.contrasts.append(
name="H2O Down",
data="H2O_dn",
background="Background 3",
bulk_in="Silicon",
bulk_out="H2O",
scalefactor="Scalefactor 3",
resolution="Resolution 1",
resample=True,
model=["DPPC absorption"],
)
problem.contrasts.append(
name="H2O Up",
data="H2O_up",
background="Background 4",
bulk_in="Silicon",
bulk_out="H2O",
scalefactor="Scalefactor 4",
resolution="Resolution 1",
resample=True,
model=["DPPC absorption"],
)
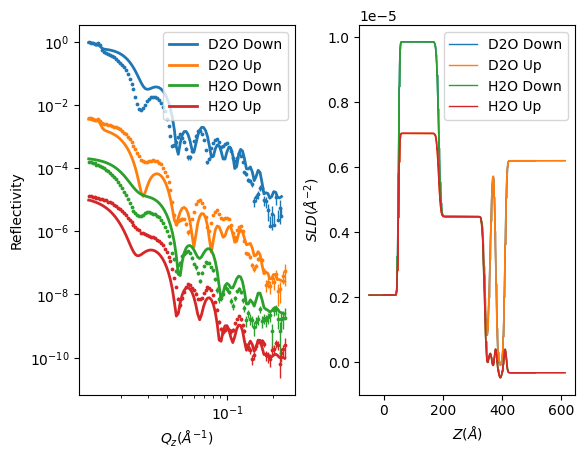
Now run RAT and plot the results.
[10]:
controls = RAT.Controls(parallel="contrasts", resampleMinAngle=0.9, resampleNPoints=150.0)
problem, results = RAT.run(problem, controls)
RAT.plotting.plot_ref_sld(problem, results)
Starting RAT ───────────────────────────────────────────────────────────────────────────────────────────────────────────
Elapsed time is 0.065 seconds
Finished RAT ───────────────────────────────────────────────────────────────────────────────────────────────────────────