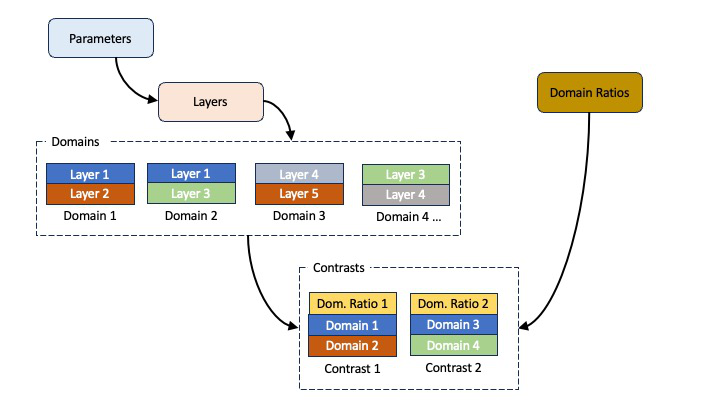
Analysing data containing domains using standard layers models is done in a similar way to a normal standard layers model, but with a couple of
additional steps.
A normal standard layers model defines parameters, which are grouped into layers, and then these are arranged into contrasts. For sample containing domains, the
layers are first grouped into domain contrasts, which are just groupings of layers with none of the additional parameters of a contrast (such as resolutions
etc). Then, the actual experimental contrasts are built in the usual way, but with the model set as any two of the domains.
To control the ratio between the domains, we can add domain ratios as described in Standard Layers Domains. And each contrast has an additional field
for domain ratios which is set as one of the values from this block.
This example can be run as a script or interactively using the instructions below.
Domains Samples Using Standard Layers
Domains Standard Layers projects proceed in much the same way as a normal SL problem, except that there is an additional grouping step between layers an contrasts.
Layers are grouped into 'Domain Contrasts'. The model for the actual experimental contrast is built from these domains rather than from layers. Thhere are exactly two domains for each contrast, with the the ratio of them controlled by a fittable 'domain ratio' parameter.
In this we will set up a simple example of a simulated system consisting of two layered domains to illustrate this process.
Start by making the project, specifying that this is a domains model...
problem = createProject(calcType="domains");
Define the parameters we need to define our two domains...
{'L1 thick', 5, 20, 60, true };
{'L1 SLD', 3e-6, 4.1e-6, 5e-6, false };
{'L1 rough' 2, 5, 20, true };
{'L1 Hydr' 10, 20, 30, true };
{'L2 thick', 5, 60, 100, true };
{'L2 SLD', 2.1e-6, 3e-6, 5e-6, false };
{'L2 rough' 2, 5, 20, true };
{'L2 Hydr' 10, 20, 30, true };
{'L3 thick', 5, 200, 300, true };
{'L3 SLD', 3e-6, 7e-6, 8e-6, false };
{'L3 rough' 2, 5, 20, true };
{'L3 Hydr' 10, 20, 30, true };
problem.addParameterGroup(Parameters);
..now group these into layers as usual....
Layer1 = {'Layer 1',... % Name of the layer
'L1 thick',... % Layer thickness
'L1 Rough',... % Layer roughness
'L1 Hydr',... % hydration (precent)
Layer2 = {'Layer 2',... % Name of the layer
'L2 thick',... % Layer thickness
'L2 Rough',... % Layer roughness
'L2 Hydr',... % hydration (precent)
Layer3 = {'Layer 3',... % Name of the layer
'L2 thick',... % Layer thickness
'L2 Rough',... % Layer roughness
'L2 Hydr',... % hydration (precent)
problem.addLayerGroup({Layer1, Layer2, Layer3});
If we look at the project, there are two extra groups as compared to a normal standard layers - Domain Contrasts and Domain Ratios...
disp(problem)
modelType: 'standard layers'
experimentName: ''
geometry: 'air/substrate'
Parameters: ----------------------------------------------------------------------------------------------
p Name Min Value Max Fit?
__ _____________________ _______ _______ _____ _____
1 "Substrate Roughness" 1 3 5 true
2 "L1 thick" 5 20 60 true
3 "L1 SLD" 3e-06 4.1e-06 5e-06 false
4 "L1 rough" 2 5 20 true
5 "L1 Hydr" 10 20 30 true
6 "L2 thick" 5 60 100 true
7 "L2 SLD" 2.1e-06 3e-06 5e-06 false
8 "L2 rough" 2 5 20 true
9 "L2 Hydr" 10 20 30 true
10 "L3 thick" 5 200 300 true
11 "L3 SLD" 3e-06 7e-06 8e-06 false
12 "L3 rough" 2 5 20 true
13 "L3 Hydr" 10 20 30 true
Bulk In: --------------------------------------------------------------------------------------------------
p Name Min Value Max Fit?
_ _________ ___ _____ ___ _____
1 "SLD Air" 0 0 0 false
Bulk Out: -------------------------------------------------------------------------------------------------
p Name Min Value Max Fit?
_ _________ _______ ________ ________ _____
1 "SLD D2O" 6.2e-06 6.35e-06 6.35e-06 false
Scalefactors: -------------------------------------------------------------------------------------------------
p Name Min Value Max Fit?
_ _______________ ____ _____ ____ _____
1 "Scalefactor 1" 0.02 0.23 0.25 false
Domain Ratios: -----------------------------------------------------------------------------------------------
p Name Min Value Max Fit?
_ ________________ ___ _____ ___ _____
1 "Domain Ratio 1" 0.4 0.5 0.6 false
Backgrounds: -----------------------------------------------------------------------------------------------
(a) Background Parameters:
p Name Min Value Max Fit?
_ ____________________ _____ _____ _____ _____
1 "Background Param 1" 1e-07 1e-06 1e-05 false
(b) Backgrounds:
p Name Type Source Value 1 Value 2 Value 3 Value 4 Value 5
_ ______________ __________ ____________________ _______ _______ _______ _______ _______
1 "Background 1" "constant" "Background Param 1" "" "" "" "" ""
Resolutions: ---------------------------------------------------------------------------------------------
(a) Resolutions Parameters:
p Name Min Value Max Fit?
_ __________________ ____ _____ ____ _____
1 "Resolution par 1" 0.01 0.03 0.05 false
(b) Resolutions:
p Name Type Source Value 1 Value 2 Value 3 Value 4 Value 5
_ ______________ __________ __________________ _______ _______ _______ _______ _______
1 "Resolution 1" "constant" "Resolution par 1" "" "" "" "" ""
Layers: --------------------------------------------------------------------------------------------------
p Name Thickness SLD Roughness Hydration Hydrate with
_ _________ __________ ________ __________ _________ ____________
1 "Layer 1" "L1 thick" "L1 SLD" "L1 rough" "L1 Hydr" "bulk out"
2 "Layer 2" "L2 thick" "L2 SLD" "L2 rough" "L2 Hydr" "bulk out"
3 "Layer 3" "L2 thick" "L2 SLD" "L2 rough" "L2 Hydr" "bulk out"
Custom Files: ------------------------------------------------------------------------------------------------------
Name Filename Function Name Language Path
____ ________ _____________ ________ ____
"" "" "" "" ""
Data: ------------------------------------------------------------------------------------------------------
Name Data Data Range Simulation Range
____________ _________ __________ _____________________
"Simulation" "No Data" "-" "[ 0.0050 , 0.7000 ]"
Domains Contrasts: -----------------------------------------------------------------------------------------------
p
_______
"Name"
"Model"
Contrasts: -----------------------------------------------------------------------------------------------
p
___________________
"Name"
"Data"
"Background"
"Background Action"
"Bulk in"
"Bulk out"
"Scalefactor"
"Resolution"
"Resample"
"Repeat Layers"
"Domain Ratio"
"Model"
Now, make a couple of Domain Contrasts, and define which layers make up which domains....
problem.addDomainContrast('name', 'Domain1', 'model', 'Layer 1');
problem.addDomainContrast('name', 'Domain2', 'model', {'Layer 2','Layer 3'});
Now make a contrast as with standard models, but this time also including the default domain ratio ("Domain Ratio 1). To set the model for our contrast, each physical contrast must have exactly two domain contrasts.
problem.addContrast('name','Domain Test',...
'background', 'Background 1',...
'resolution', 'Resolution 1',...
'scalefactor', 'Scalefactor 1',...
'domainRatio', 'Domain Ratio 1',...
'model', {'Domain1', 'Domain2'});
Now we can run our simulation as usual, and plot the results....
% Make a controls class....
controls = controlsClass();
% Send everything to RAT....
[problem,results] = RAT(problem,controls);
Starting RAT ______________________________________________________________________________________________
Elapsed time is 0.018835 seconds.
Finished RAT ______________________________________________________________________________________________
plotRefSLD(problem,results);