Note
To get the output project and results from this example in your Python session, run:
from ratapi.examples import DSPC_custom_layers
project, results = DSPC_custom_layers()
[1]:
import pathlib
import numpy as np
from IPython.display import Code
import ratapi as RAT
from ratapi.models import Parameter
Custom Layers Example for Supported DSPC layer#
Example of using Custom layers to model a DSPC supported bilayer. Start by making the project and setting it to a custom layers type:
[2]:
problem = RAT.Project(name="Orso lipid example - custom layers", model="custom layers", geometry="substrate/liquid")
problem.show_priors()
For a custom layers model, rather than being forced to define our layers as [Thick SLD Rough…. etc], we can parameterise however we like and then use a function to calculate the [d \(\rho\) \(\sigma\)] arrangement for each layer. So for example, if the volume of lipid tails are known (from the literature), then all we need is the Area per molecule, because then:
where d is the thickness and V is the volume.
Likewise, the SLD is:
as usual.
In this folder there is a pre-prepared Python custom model for a DSPC on a Silicon substrate. We can display it here to see what we mean:
[3]:
Code(filename='custom_bilayer_DSPC.py', language='python')
[3]:
"""A custom layer model for a DSPC supported bilayer."""
import numpy as np
def custom_bilayer_DSPC(params, bulk_in, bulk_out, contrast):
"""Calculate layer parameters for a DSPC supported bilayer.
This file accepts 3 vectors containing the values for params, bulk in and bulk out.
The final parameter is an index of the contrast being calculated.
The function should output a matrix of layer values, in the form...
Output = [thick 1, SLD 1, Rough 1, Percent Hydration 1, Hydrate how 1
....
thick n, SLD n, Rough n, Percent Hydration n, Hydration how n]
The "hydrate how" parameter decides if the layer is hydrated with Bulk out or Bulk in phases.
Set to 1 for Bulk out, zero for Bulk in.
Alternatively, leave out hydration and just return...
Output = [thick 1, SLD 1, Rough 1,
....
thick n, SLD n, Rough n]
The second output parameter should be the substrate roughness.
"""
# Note - The first contrast number is 1 (not 0) so be careful if you use
# this variable for array indexing.
sub_rough = params[0]
oxide_thick = params[1]
oxide_hydration = params[2]
lipidAPM = params[3]
headHydration = params[4]
bilayerHydration = params[5]
bilayerRough = params[6]
waterThick = params[7]
# We have a constant SLD for the bilayer
oxide_SLD = 3.41e-6
# Now make the lipid layers
# Use known lipid volume and compositions to make the layers
# define all the neutron b's.
bc = 0.6646e-4 # Carbon
bo = 0.5843e-4 # Oxygen
bh = -0.3739e-4 # Hydrogen
bp = 0.513e-4 # Phosphorus
bn = 0.936e-4 # Nitrogen
# Now make the lipid groups
COO = (4 * bo) + (2 * bc)
GLYC = (3 * bc) + (5 * bh)
CH3 = (2 * bc) + (6 * bh)
PO4 = (1 * bp) + (4 * bo)
CH2 = (1 * bc) + (2 * bh)
CHOL = (5 * bc) + (12 * bh) + (1 * bn)
# Group these into heads and tails:
Head = CHOL + PO4 + GLYC + COO
Tails = (34 * CH2) + (2 * CH3)
# We need volumes for each. Use literature values:
vHead = 319
vTail = 782
# We use the volumes to calculate the SLDs
SLDhead = Head / vHead
SLDtail = Tails / vTail
# We calculate the layer thickness' from the volumes and the APM
headThick = vHead / lipidAPM
tailThick = vTail / lipidAPM
# Manually deal with hydration for layers in this example.
oxSLD = (oxide_hydration * bulk_out[contrast - 1]) + ((1 - oxide_hydration) * oxide_SLD)
headSLD = (headHydration * bulk_out[contrast - 1]) + ((1 - headHydration) * SLDhead)
tailSLD = (bilayerHydration * bulk_out[contrast - 1]) + ((1 - bilayerHydration) * SLDtail)
# Make the layers
oxide = [oxide_thick, oxSLD, sub_rough]
water = [waterThick, bulk_out[contrast - 1], bilayerRough]
head = [headThick, headSLD, bilayerRough]
tail = [tailThick, tailSLD, bilayerRough]
output = np.array([oxide, water, head, tail, tail, head])
return output, sub_rough
We need to add the parameters we are going to need to define the model (note that Substrate Roughness always exists as parameter 0 as before, and that we are setting a Gaussian prior on the Head Hydration here).
[4]:
parameter_list = [
Parameter(name="Oxide Thickness", min=5.0, value=20.0, max=60.0, fit=True),
Parameter(name="Oxide Hydration", min=0.0, value=0.2, max=0.5, fit=True),
Parameter(name="Lipid APM", min=45.0, value=55.0, max=65.0, fit=True),
Parameter(name="Head Hydration", min=0.0, value=0.2, max=0.5, fit=True, prior_type='gaussian', mu=0.3, sigma=0.03),
Parameter(name="Bilayer Hydration", min=0.0, value=0.1, max=0.2, fit=True),
Parameter(name="Bilayer Roughness", min=2.0, value=4.0, max=8.0, fit=True),
Parameter(name="Water Thickness", min=0.0, value=2.0, max=10.0, fit=True)
]
problem.parameters.extend(parameter_list)
problem.parameters.set_fields(0, min=1.0, max=10.0)
Need to add the relevant Bulk SLD’s. Change the bulk in from air to silicon, and add two additional water contrasts:
[5]:
# Change the bulk in from air to silicon:
problem.bulk_in.set_fields(0, name="Silicon", min=2.07e-6, value=2.073e-6, max=2.08e-6, fit=False)
problem.bulk_out.append(name="SLD SMW", min=1.0e-6, value=2.073e-6, max=3.0e-6, fit=True)
problem.bulk_out.append(name="SLD H2O", min=-0.6e-6, value=-0.56e-6, max=-0.3e-6, fit=True)
problem.bulk_out.set_fields(0, min=5.0e-6, fit=True)
Now add the datafiles. We have three datasets we need to consider - the bilayer against D2O, Silicon Matched water and H2O. Load these datafiles in and put them in the data block:
[6]:
# Read in the datafiles
data_path = pathlib.Path("../data")
D2O_data = np.loadtxt(data_path / "c_PLP0016596.dat", delimiter=",")
SMW_data = np.loadtxt(data_path / "c_PLP0016601.dat", delimiter=",")
H2O_data = np.loadtxt(data_path / "c_PLP0016607.dat", delimiter=",")
# Add the data to the project - note this data has a resolution 4th column
problem.data.append(name="Bilayer / D2O", data=D2O_data, data_range=[0.013, 0.37])
problem.data.append(name="Bilayer / SMW", data=SMW_data, data_range=[0.013, 0.32996])
problem.data.append(name="Bilayer / H2O", data=H2O_data, data_range=[0.013, 0.33048])
Add the custom file to the project:
[7]:
problem.custom_files.append(name="DSPC Model", filename="custom_bilayer_DSPC.py", language="python", path=pathlib.Path.cwd().resolve())
Also, add the relevant background parameters - one each for each contrast:
[8]:
problem.background_parameters.set_fields(0, name="Background parameter D2O", min=1.0e-10, max=1.0e-5, value=1.0e-07, fit=True)
problem.background_parameters.append(name="Background parameter SMW", min=1.0e-10, value=1.0e-7, max=1.0e-5, fit=True)
problem.background_parameters.append(name="Background parameter H2O", min=1.0e-10, value=1.0e-7, max=1.0e-5, fit=True)
# And add the two new constant backgrounds
problem.backgrounds.append(name="Background SMW", type="constant", source="Background parameter SMW")
problem.backgrounds.append(name="Background H2O", type="constant", source="Background parameter H2O")
# And edit the other one
problem.backgrounds.set_fields(0, name="Background D2O", source="Background parameter D2O")
# Finally modify some of the other parameters to be more suitable values for a solid / liquid experiment
problem.scalefactors.set_fields(0, value=1.0, min=0.5, max=2.0, fit=True)
We need to use the data resolution (i.e. the fourth column of our datafiles). Do do this, we need to add a ‘Data’ resolution object to our resolutions table
[9]:
problem.resolutions.append(name="Data Resolution", type="data")
Now add the three contrasts as before:
[10]:
problem.contrasts.append(
name="Bilayer / D2O",
background="Background D2O",
resolution="Data Resolution",
scalefactor="Scalefactor 1",
bulk_out="SLD D2O",
bulk_in="Silicon",
data="Bilayer / D2O",
model=["DSPC Model"],
)
problem.contrasts.append(
name="Bilayer / SMW",
background="Background SMW",
resolution="Data Resolution",
scalefactor="Scalefactor 1",
bulk_out="SLD SMW",
bulk_in="Silicon",
data="Bilayer / SMW",
model=["DSPC Model"],
)
problem.contrasts.append(
name="Bilayer / H2O",
background="Background H2O",
resolution="Data Resolution",
scalefactor="Scalefactor 1",
bulk_out="SLD H2O",
bulk_in="Silicon",
data="Bilayer / H2O",
model=["DSPC Model"],
)
Note that the model is simply the custom file we’ve just added to the project.
Look at the complete model definition before sending it to RAT:
[11]:
print(problem)
Name: ----------------------------------------------------------------------------------------------
Orso lipid example - custom layers
Calculation: ---------------------------------------------------------------------------------------
normal
Model: ---------------------------------------------------------------------------------------------
custom layers
Geometry: ------------------------------------------------------------------------------------------
substrate/liquid
Parameters: ----------------------------------------------------------------------------------------
+-------+---------------------+------+-------+------+------+------------+
| index | name | min | value | max | fit | prior type |
+-------+---------------------+------+-------+------+------+------------+
| 0 | Substrate Roughness | 1.0 | 3.0 | 10.0 | True | uniform |
| 1 | Oxide Thickness | 5.0 | 20.0 | 60.0 | True | uniform |
| 2 | Oxide Hydration | 0.0 | 0.2 | 0.5 | True | uniform |
| 3 | Lipid APM | 45.0 | 55.0 | 65.0 | True | uniform |
| 4 | Head Hydration | 0.0 | 0.2 | 0.5 | True | gaussian |
| 5 | Bilayer Hydration | 0.0 | 0.1 | 0.2 | True | uniform |
| 6 | Bilayer Roughness | 2.0 | 4.0 | 8.0 | True | uniform |
| 7 | Water Thickness | 0.0 | 2.0 | 10.0 | True | uniform |
+-------+---------------------+------+-------+------+------+------------+
Bulk In: -------------------------------------------------------------------------------------------
+-------+---------+----------+-----------+----------+-------+------------+
| index | name | min | value | max | fit | prior type |
+-------+---------+----------+-----------+----------+-------+------------+
| 0 | Silicon | 2.07e-06 | 2.073e-06 | 2.08e-06 | False | uniform |
+-------+---------+----------+-----------+----------+-------+------------+
Bulk Out: ------------------------------------------------------------------------------------------
+-------+---------+--------+-----------+----------+------+------------+
| index | name | min | value | max | fit | prior type |
+-------+---------+--------+-----------+----------+------+------------+
| 0 | SLD D2O | 5e-06 | 6.35e-06 | 6.35e-06 | True | uniform |
| 1 | SLD SMW | 1e-06 | 2.073e-06 | 3e-06 | True | uniform |
| 2 | SLD H2O | -6e-07 | -5.6e-07 | -3e-07 | True | uniform |
+-------+---------+--------+-----------+----------+------+------------+
Scalefactors: --------------------------------------------------------------------------------------
+-------+---------------+-----+-------+-----+------+------------+
| index | name | min | value | max | fit | prior type |
+-------+---------------+-----+-------+-----+------+------------+
| 0 | Scalefactor 1 | 0.5 | 1.0 | 2.0 | True | uniform |
+-------+---------------+-----+-------+-----+------+------------+
Background Parameters: -----------------------------------------------------------------------------
+-------+--------------------------+-------+-------+-------+------+------------+
| index | name | min | value | max | fit | prior type |
+-------+--------------------------+-------+-------+-------+------+------------+
| 0 | Background parameter D2O | 1e-10 | 1e-07 | 1e-05 | True | uniform |
| 1 | Background parameter SMW | 1e-10 | 1e-07 | 1e-05 | True | uniform |
| 2 | Background parameter H2O | 1e-10 | 1e-07 | 1e-05 | True | uniform |
+-------+--------------------------+-------+-------+-------+------+------------+
Backgrounds: ---------------------------------------------------------------------------------------
+-------+----------------+----------+--------------------------+
| index | name | type | source |
+-------+----------------+----------+--------------------------+
| 0 | Background D2O | constant | Background parameter D2O |
| 1 | Background SMW | constant | Background parameter SMW |
| 2 | Background H2O | constant | Background parameter H2O |
+-------+----------------+----------+--------------------------+
Resolution Parameters: -----------------------------------------------------------------------------
+-------+--------------------+------+-------+------+-------+------------+
| index | name | min | value | max | fit | prior type |
+-------+--------------------+------+-------+------+-------+------------+
| 0 | Resolution Param 1 | 0.01 | 0.03 | 0.05 | False | uniform |
+-------+--------------------+------+-------+------+-------+------------+
Resolutions: ---------------------------------------------------------------------------------------
+-------+-----------------+----------+--------------------+---------+
| index | name | type | source | value 1 |
+-------+-----------------+----------+--------------------+---------+
| 0 | Resolution 1 | constant | Resolution Param 1 | |
| 1 | Data Resolution | data | | |
+-------+-----------------+----------+--------------------+---------+
Custom Files: --------------------------------------------------------------------------------------
+-------+------------+------------------------+---------------------+----------+------------------------------------------------------------------------------+
| index | name | filename | function name | language | path |
+-------+------------+------------------------+---------------------+----------+------------------------------------------------------------------------------+
| 0 | DSPC Model | custom_bilayer_DSPC.py | custom_bilayer_DSPC | python | /opt/actions-runner/_work/RAT-Docs/RAT-Docs/source/python_examples/notebooks |
+-------+------------+------------------------+---------------------+----------+------------------------------------------------------------------------------+
Data: ----------------------------------------------------------------------------------------------
+-------+---------------+-----------------------+------------------+----------------------+
| index | name | data | data range | simulation range |
+-------+---------------+-----------------------+------------------+----------------------+
| 0 | Simulation | [] | [] | [0.005, 0.7] |
| 1 | Bilayer / D2O | Data array: [146 x 4] | [0.013, 0.37] | [0.0057118, 0.39606] |
| 2 | Bilayer / SMW | Data array: [97 x 4] | [0.013, 0.32996] | [0.0076029, 0.32996] |
| 3 | Bilayer / H2O | Data array: [104 x 4] | [0.013, 0.33048] | [0.0063374, 0.33048] |
+-------+---------------+-----------------------+------------------+----------------------+
Contrasts: -----------------------------------------------------------------------------------------
+-------+---------------+---------------+----------------+-------------------+---------+----------+---------------+-----------------+----------+---------------+------------+
| index | name | data | background | background action | bulk in | bulk out | scalefactor | resolution | resample | repeat layers | model |
+-------+---------------+---------------+----------------+-------------------+---------+----------+---------------+-----------------+----------+---------------+------------+
| 0 | Bilayer / D2O | Bilayer / D2O | Background D2O | add | Silicon | SLD D2O | Scalefactor 1 | Data Resolution | False | 1 | DSPC Model |
| 1 | Bilayer / SMW | Bilayer / SMW | Background SMW | add | Silicon | SLD SMW | Scalefactor 1 | Data Resolution | False | 1 | DSPC Model |
| 2 | Bilayer / H2O | Bilayer / H2O | Background H2O | add | Silicon | SLD H2O | Scalefactor 1 | Data Resolution | False | 1 | DSPC Model |
+-------+---------------+---------------+----------------+-------------------+---------+----------+---------------+-----------------+----------+---------------+------------+
To run it, we need to make a controls block
[12]:
controls = RAT.Controls()
print(controls)
+---------------------+-----------+
| Property | Value |
+---------------------+-----------+
| procedure | calculate |
| parallel | single |
| numSimulationPoints | 500 |
| resampleMinAngle | 0.9 |
| resampleNPoints | 50 |
| display | iter |
+---------------------+-----------+
… and send this to RAT
[13]:
problem, results = RAT.run(problem, controls)
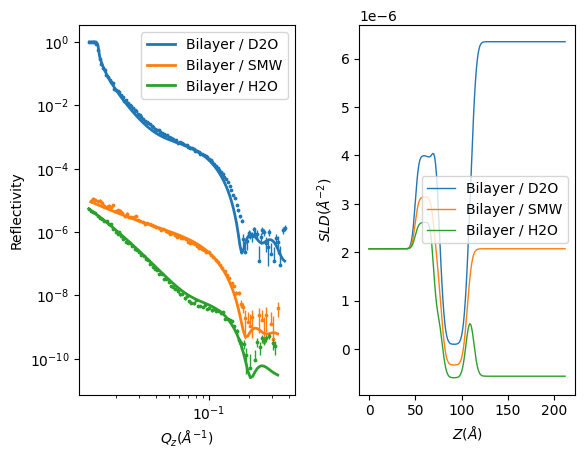
RAT.plotting.plot_ref_sld(problem, results)
Starting RAT ───────────────────────────────────────────────────────────────────────────────────────────────────────────
Elapsed time is 0.013 seconds
Finished RAT ───────────────────────────────────────────────────────────────────────────────────────────────────────────